Butterfly eyespots evolved via cooption of an ancestral gene-regulatory network that also patterns antennae, legs, and wings | PNAS

Genomic evidence for homoploid hybrid speciation in a marine mammal apex predator | Science Advances

Multiplex-GAM: genome-wide identification of chromatin contacts yields insights overlooked by Hi-C | Nature Methods
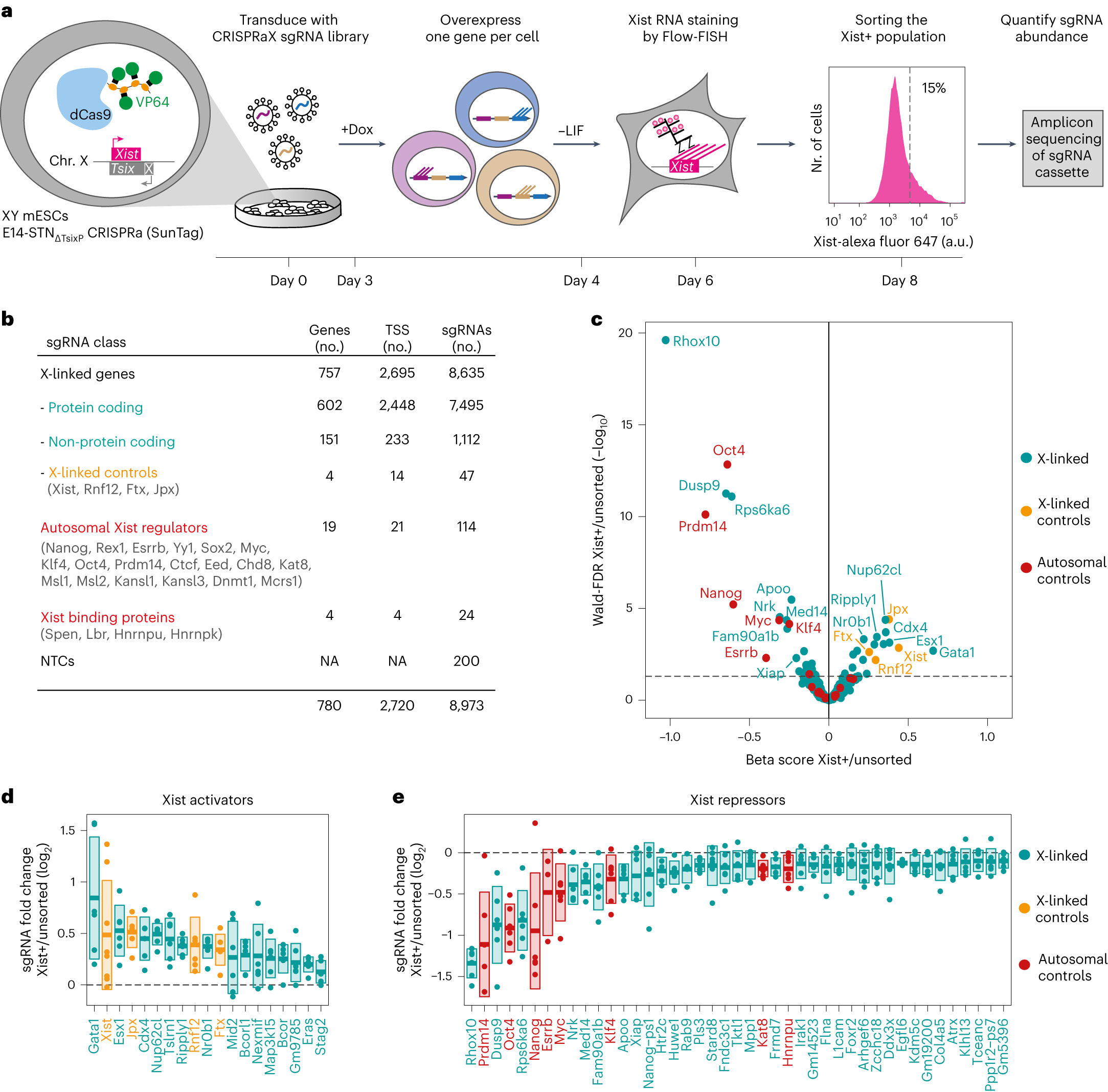
GATA transcription factors drive initial Xist upregulation after fertilization through direct activation of long-range enhancers | Nature Cell Biology
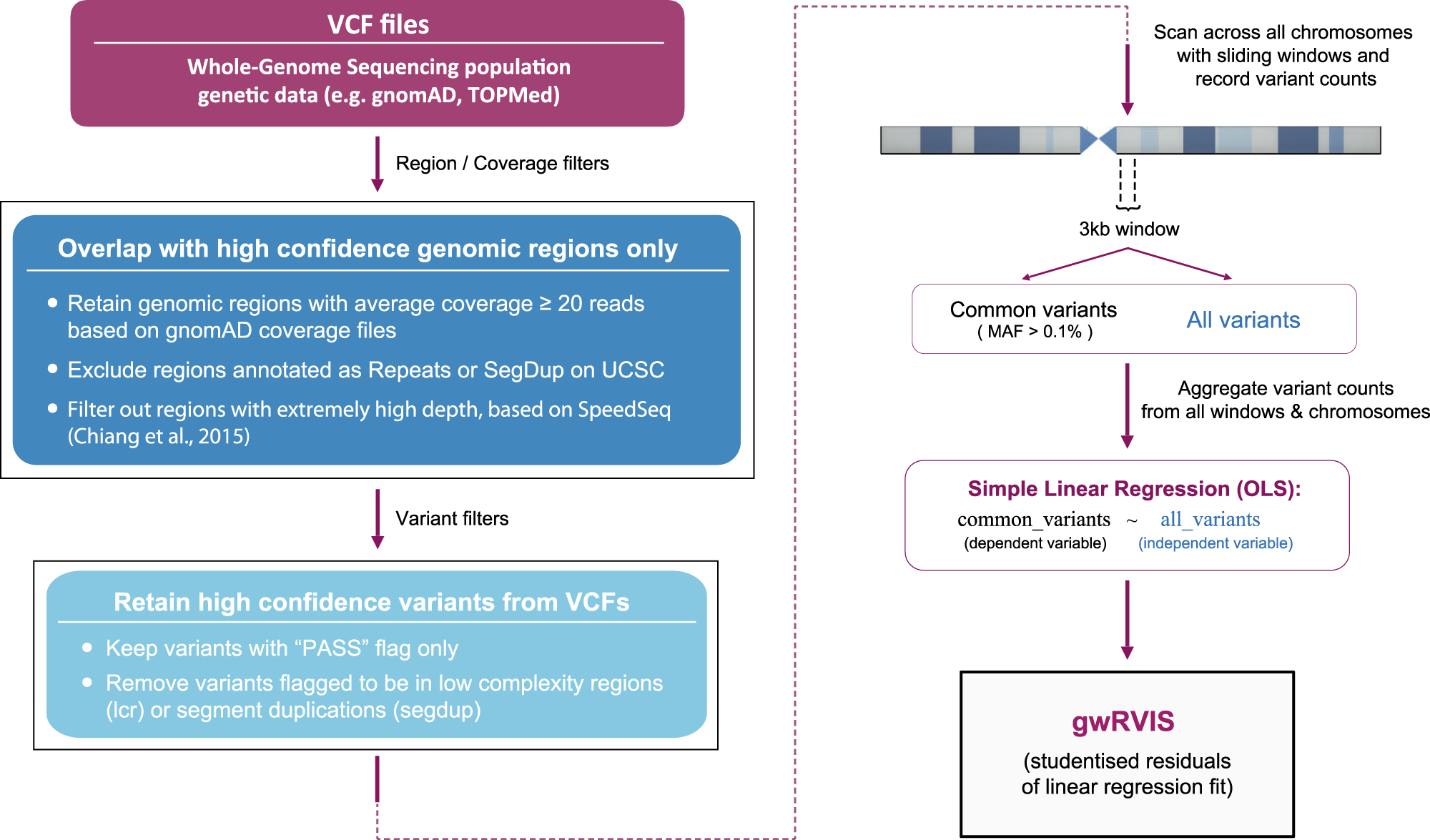
Prioritizing non-coding regions based on human genomic constraint and sequence context with deep learning | Nature Communications

Highly sensitive single-cell chromatin accessibility assay and transcriptome coassay with METATAC | PNAS

exRNA-eCLIP intersection analysis reveals a map of extracellular RNA binding proteins and associated RNAs across major human biofluids and carriers - ScienceDirect